Dislocation Defect Analyzer
The Dislocation Defect analyzer uses a trained AI model to identify and classify defects found in images of a scan. This article provides an overview about dislocation defects and how the Dislocation Defect analyzer works in nSpec.
Overview
Dislocations are line defects that exist in the crystal structure of metals. More information about dislocations can be found here.
The goal of the Dislocation Defect analyzer in nSpec is to find and classify dislocation defects on etched silicon carbide (SiC) substrates. Specifically, the Dislocation Defect analyzer does the following to find and classify dislocations in a provided image:
Find irregular shapes and patterns
Determine if that shape or pattern is a defect
Classify that defect using a trained AI model
Historically, we have had to generate entirely unique builds of nSpec to deploy different flavors of the Dislocation Analyzer at customer sites. This has caused a problematic divergence of features and functionalities available to these customers over time, sometimes even across different nSpec systems at the same customer. This is the most important step towards enabling these customers to get back on the main branch of software improvements, while also dramatically improving our core Dislocation Defect Analyzer. This work also sets the stage for easier customer-specific customization of our dislocation analyzer moving forward.
For complete instructions on how to train and configure the new Dislocation Analyzer, please reach out to us at support@nanotronics.ai
Dislocation Classification Types
This article uses a sample set of dislocation classifications:
These are only some types of dislocations - there are many more. The Dislocation Defect analyzer can classify defects using the provided sample classifications above or use another set of classifications. Furthermore, defects found using the Dislocation Defect analyzer can be considered a chain if there is more than one defect overlapping another.
Classification Functionality
The classification of defects is done by a utility that uses a machine learning technology known as Support Vector Machine (SVM). SVMs are supervised learning models that train using a set of pre-classified samples. The Dislocation Defect analyzer uses this trained model perform its classifications of found defects.
Configuration File - nJson
The user now has the ability to select between multiple analyzer configurations when creating analysis parameter groups. For all analyzers, these configuration files, known as nJson files, can affect the following parameter properties:
Visibility in the dialog
Default process value
Low/High limits
nJson is a UTF-8-encoded and JSON-formatted file type used by nSpec for analysis and by the SVM training utility for training. The following is a sample nJson file:
{
"parameters":
[
{"task": "hide", "name": "(DEBUG ONLY) Export Defects", "value": "FALSE"},
{"task": "modify", "ParameterGUID": "{95803C07-74CD-44D2-BE5D-6C498059EA7D}","values": {"LoLim": "0.1","HiLim": "0.9"}}
],
"classifications":
[
{"class": "BPD", "sub_class": "BPD" "is_chain": "FALSE"},
{"name": "TED", "is_chain": "FALSE"},
{"name": "TSD", "is_chain": "FALSE"},
{"name": "Chain", "is_chain": "TRUE"},
{"name": "Other", "is_chain": "FALSE"}
],
"composite2component":
[
{"composite": "Chain", "component": "TED" }
],
"otherClass" : "Other",
"chainClass": "Chain",
"autoThresholdMethod": "KMeans",
"model": "eFullPixel",
"thresholdFillInValue": 255,
"epsilon": 0.0,
"SVM_Cutoffs":
[
{"class": "BPD", "value": 10.0},
{"class": "TED", "value": 1070000000.0},
{"class": "TSD", "value": 1077000000.0}
]
}Customers are not expected to configure nJson files. Contact support@nanotronics.ai for assistance in creating dislocation classification models.
Renaming Classifications
The SVM trainer does not recognize the actual names of classifications in the classifications property, only their positions within the classifications property's array. Therefore, it is possible to rename classifications on the fly by changing their names in the nJson file (no retraining required).
Automatically Selecting Models
For the Dislocation Defect analyzer, a useful ability of the parameters property is to auto-select models. There are two Dislocation Defect analyzer parameters that can be used to specify a model:
Parameter | Usage |
| Set this to the absolute path of the target model. |
| Set this to the name of the target model within |
If you wish to auto-select a model, you should:
Hide
Configuration File Pathanalysis parameter and set itsvalueproperty to"".Hide
configFileNameanalysis parameter (should always be hidden) and set itsvalueproperty to the target model file (i.e.,DislocationTestModel.xml).Ensure target model file is inside
C:\Nanotronics Imaging\ProgramData\ConfigFiles\Analysis\Dislocation\ModelFiles.
Creating a Parameter Group
The Dislocation Defect analyzer is unique among nSpec analyzers in that it requires a nJson file. This requirement makes the Dislocation Defect analyzer parameters and Analysis Parameters dialog configured a little differently then other analyzers. The workflow for creating a Dislocation Defect analyzer is documented in this section.
1 - Place Proper Files in ConfigFiles Folder
To successfully use the Dislocation Analyzer in nSpec, you will need the following:
nJson file
SVM model (.xml file)
1.1 - Navigate to Dislocation Config Folder
Navigate to the C:\Nanotronics Imaging\ProgramData\ConfigFiles\Analysis\Dislocation directory. This directory is where all your Dislocation Defect analyzer related files go.
1.2 - Copy Your nJson
In the current directory, copy your desired nJson file. Your directory should look like the following:

At this point, if you do not need it, archive the Default.nJson to a backup location. Keeping this file in this location is ok, but will convolute the parameter group creation process and will create extra dialogs for users to click through.
1.3 - Copy Your SVM Model
In the ModelFiles directory, copy your desired SVM model (.xml). Your ModelFiles directory should look like the following:
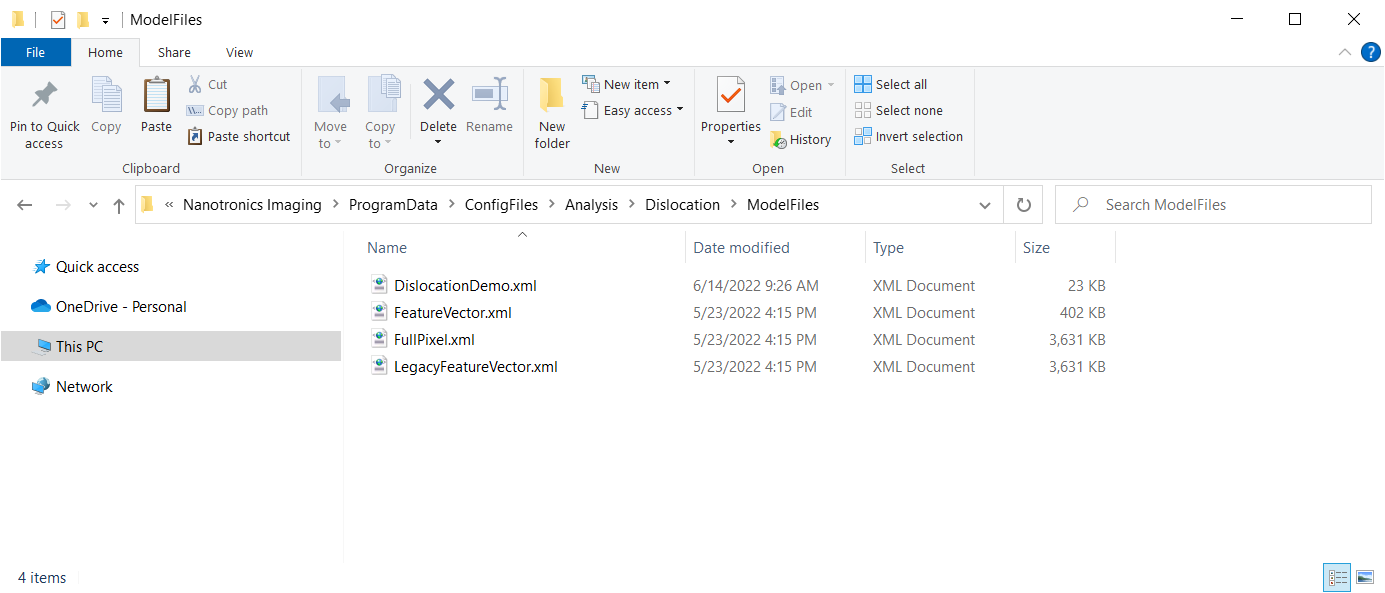
You can leave the other SVM models (.xml) alone. They will not affect anything.
2 - Open Analysis Parameters Dialog
Open the Analysis Parameters dialog for the Dislocation Defect analyzer.
2.1 - Navigate to Analysis Parameters Dialog
In nSpec, navigate to nSpec Main View > Analysis > Edit Parameters…
If the Dislocation Defect analyzer is not pre-selected, select the Dislocation Defect Analyzer in the Analysis combo box at the top of the Analysis Parameters dialog.
2.2 - Select nJson File
NOTE: If you deleted the Default.nJson file (and therefore only have one nJson in the C:\Nanotronics Imaging\ProgramData\ConfigFiles\Analysis\Dislocation directory), you may go to step 2.3.
If you have more than one nJson file in the C:\Nanotronics Imaging\ProgramData\ConfigFiles\Analysis\Dislocation directory, you will get the following dialog:
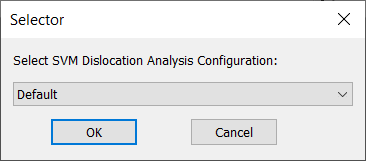
Please select your desired nJson using the combo box and press OK.
2.3 - Understand The Save Button
When you first see the Analysis Parameters dialog, you will notice that the Save button is enabled. Furthermore, if you select another parameter group within the Dislocation Defect analyzer or select another analyzer, you will get a warning that your parameter group has not been saved.
The reason for this that the Dislocation Defect analyzer has two completely hidden parameters that automatically get filled in when you open the Analysis Parameters dialog:
JsonConfigTextJsonConfigFilePath
Just be aware that this functionality happens every single time you entire this dialog.
3 - Set Parameters
At this point, the Dislocation Defect analyzer is just like every other analyzer in terms of setting parameters.
Dislocation Defect Analyzer Parameters
Below is a summary of all the available Dislocation Defect analyzer parameters.
Parameter | Description | Advice |
|---|---|---|
Configuration File Path | Absolute path to model file |
|
configFileName | Name of model file in ModelFiles folder |
|
Calculate Exclusion Info | In the exported CSV report, state whether each defect is in the included wafer area. | |
Look for Non-Chain Dislocation defects | Look for non-chain dislocation defects | |
Export CSV Containing Defect Info | Export a report containing defect data to a CSV. |
|
Look for Chain Defects | Look for chain defects. Turning this option means corner masking will be performed. | |
Minimum Chain Eccentricity | Minimum chain eccentricity | |
Minimum Chain Area | (Micrometers Squared) Minimum chain area. | |
Minimum Dislocation Defect Area | (Micrometers Squared) Minimum chain area. | |
Use Automatic Thresholding. | Use automatic thresholding. | |
Manual Threshold Value | Manual threshold value. Only relevant if automatic thresholding is off. | |
Use Automatic Corner thresholding | Use automatic thresholding when looking for corners. Only relevant if doing corner masking. | |
Manual Corner Threshold | Manual Corner Threshold. Only relevant if automatic corner thresholding is off. | |
Force Corner Masking | Force corner masking regardless of whether or not the analyzer is looking for chains. | |
Minimum Corner Side Length | (Micrometers) Minimum side length of the stage as it appears on images of the wafer's edge. | |
Calculate chain area ratio data | Export data relating the area of chains to the area of component defects. | |
Chain area overlap percentage | The overlap of component defects that create chains. Only used in chain area ratio calculations. | |
Exclude Other defects | Exclude (ignore) defects that classified as Other. | |
Export JSON | Set to TRUE to export analysis results to JSON file. Set to FALSE to disable. |
Export To CSV
All historical versions of the Dislocation Defect analyzer had an option to export to CSV. The output CSV of each of these analyzers had the same column structure but different parameters.
Under C:\Nanotronics Imaging\ProgramData\ConfigFiles\Analysis\Dislocation\Queries, there is a SQL file that is used to prepare the data for the Export CSV Containing Defect Info analysis parameter. This file query execution depends on temporary view that is created just before launching the analysis. Do not touch, modify, remove, rename, or do anything with this file.
