Custom Export File Formatting with String Templates
Overview
Users can create custom formatted export files in Custom Exporter using user-defined nfmt (Nano format) files.
This guide assume familiarity with Python f-string formatting. nfmt is an implementation of {fmt} C++ library. We’ve developed a custom syntax-highlighting extension for VS Code to aid in creating nfmt files. Contact support@nanotronics.ai for access to this custom VS extension.
Custom Exporter Usage
To use the custom string template formatter in Custom Exporter, you must select UserDefined as the Export Type.
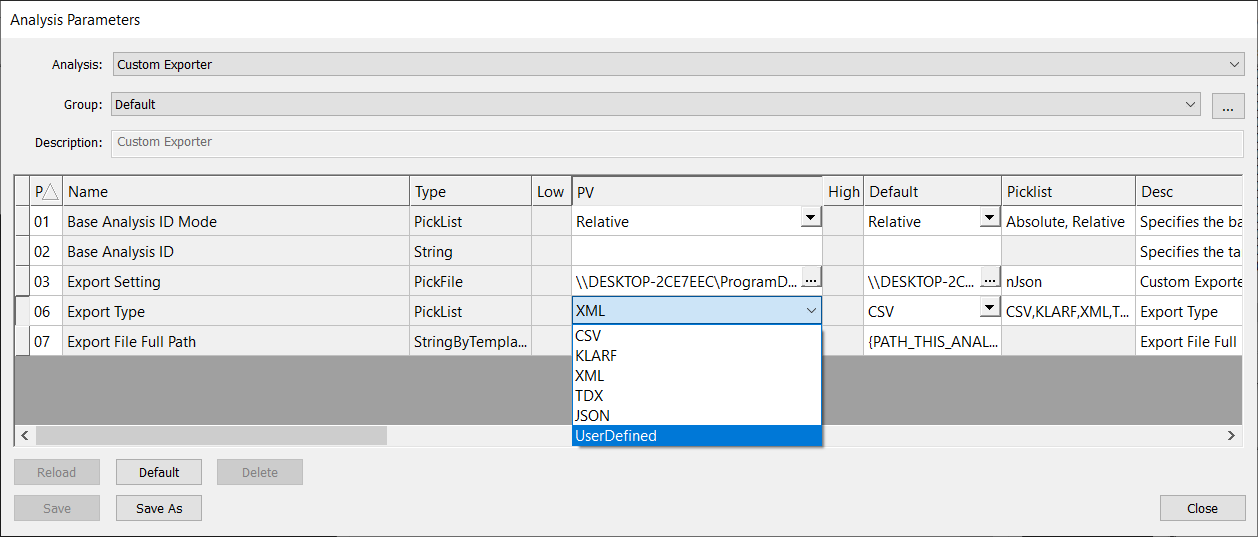
When selecting UserDefined export type, you must provide a nfmt file to format your export. You must point to this nfmt file using the field CustomTypeFile in the Export Setting nJson file.
Below is an example nJson file. Note that the CustomTypeFile is nanofmt.nfmt and the CustomTypeExtension is txt.
{
"Custom Exporter": {
"inputTablesNames": "src",
"CustomTypeExtension": "txt",
"CustomTypeFile": "nanofmt.nfmt",
"Views" : [
{
"Title": "srcScanID",
"Query": "SELECT ScanID FROM vwAnalysis WHERE AnalysisID = (SELECT AnalysisID FROM src LIMIT 1)"
},
{
"Title": "vwSrcScan",
"Query": "SELECT ScanID, Timestamp As StartTime, EndTime, (SELECT COUNT(*) FROM vwImages) AS NumImages , (SELECT PropertyData / 1000 FROM ScanProperties WHERE PropertyName = 'Scan Min X Microns') AS 'X_mm', (SELECT PropertyData / 1000 FROM ScanProperties WHERE PropertyName = 'Scan Min Y Microns') AS 'Y_mm', (SELECT PropertyData / 1000 FROM ScanProperties WHERE PropertyName = 'Scan Width Microns') AS 'Width_mm', (SELECT PropertyData / 1000 FROM ScanProperties WHERE PropertyName = 'Scan Height Microns') AS 'Height_mm', Result, (SELECT PropertyData FROM ScanProperties WHERE PropertyName = 'SampleID') AS SampleName FROM Scans WHERE ScanID = (SELECT ScanID FROM srcScanID)"
},
{
"Title": "vwSrcAnalysis",
"Query": "SELECT AnalysisID, AnalyzerMenuText AS Analyzer, Timestamp AS StartTime, EndTime, NumDefects, AnalyzerName, Result FROM vwAnalysis WHERE AnalysisID = (SELECT AnalysisID FROM src LIMIT 1)"
}
],
"Queries": [
{
"Title": "scans",
"Query": "SELECT * FROM Scans"
}
]
}
}Example nfmt formatting file
Below is an example nfmt file. Anything not contained in braces { } is considered literal text, and brace characters can be escaped by doubling {{ }}.
Note that within the braces, the nfmt file references and accesses the SQL database queries made in the nJson Export Setting file, e.g. scans and vsSrcScan.
User-Defined Report Sample (from Query)
=======================================
Scans ID : {scans:e{"\t"}:ScanID}
Scans StartTime : {scans:e{"\t"}:Timestamp}
Scans EndTime : {scans:e{"\t"}:EndTime}
Scans Result : {scans:e{"\t"}:Result}
{db:sql{scan, SELECT * FROM vwSrcScan}:e{"\n"}:f{
"
Defined Report Sample (from views)
=======================================
Scan ID : {scan:ScanID}
Scan StartTime : {scan:StartTime}
Scan EndTime : {scan:EndTime}
Number of Images : {scan:NumImages}
X(mm) : {scan:X_mm}
Y(mm) : {scan:Y_mm}
Width(mm) : {scan:Width_mm}
Height(mm) : {scan:Height_mm}
Scan Result : {scan:Result}
SampleName : {scan:SampleName}
{db:
sql{property,
f{"SELECT * FROM ScanProperties WHERE ScanID = {scan:ScanID}"} }
:e{"\n"}
:f{"{property:PropertyName} : {property:PropertyData}"} }
{db:sql{analysis, SELECT * FROM vwAnalysis WHERE AnalysisID = (SELECT AnalysisID
FROM src LIMIT 1)}
:e{"\n"}
:f{
"
Analyzer
--------
StartTime : {analysis:Timestamp}
EndTime : {analysis:EndTime}
Defects : {analysis:NumDefects}
AnalyzerName : {analysis:AnalyzerName}
"} }
Devices
-------
{db:
sql{device, f{"SELECT * FROM vwDevices WHERE ScanID = {scan:ScanID} ORDER BY
DeviceID"} }
:e{"\n"}
:f{
"
DeviceID : {device:DeviceID}
X : {device:X}
Y : {device:Y}
width : {device:width}
height : {device:height}
Images
------
{db:sql{image, f{"
SELECT * FROM vwImages WHERE ImageID IN
(SELECT ImageID FROM DevicesInImages WHERE DeviceID = {device:DeviceID})
ORDER BY ImageID"} }
:e{"\n"}
:f{
"
ImageID : {image:ImageID}
X : {image:X}
Y : {image:Y}
width : {image:wMicrons}
height : {image:hMicrons}
Defects
-------
DefectID X Y W H Area
{db:sql{d, f{"SELECT * FROM src WHERE ImageID = {image:ImageID} ORDER BY DefectID"}
}
::e{" {d:DefectID:<11} {d:X:<6.5g} {d:Y:<6.5g} {d:W:<6.5g} {d:H:<6.5g}
{d:Area:<9.8g}\n"
} }"
} }"
} }"
} }
Example Export File
The following export file was generated via Custom Exporter using the example nfmt file above. The data is sample data, but typically the data draws from the nSpec scan database. Only a portion of the Defects have been included for brevity.
User-Defined Report Sample (from Query)
=======================================
Scans ID : 1 2 3 4 5 6
Scans StartTime : 2024-09-08 22:45:10.624 2024-09-08 22:47:04.214 2024-09-08 22:48:05.204 2024-09-08 22:50:10.931 2024-09-08 22:51:09.385 2024-09-08 22:53:06.674
Scans EndTime : 2024-09-08 22:47:03.977 2024-09-09 12:48:39.323 2024-09-08 22:50:08.973 2024-09-09 12:55:33.344 2024-09-08 22:53:04.566 2024-09-09 13:00:17.962
Scans Result : Success Success Success Success Success Success
Defined Report Sample (from views)
=======================================
Scan ID : 6
Scan StartTime : 2024-09-08 22:53:06.674
Scan EndTime : 2024-09-09 13:00:17.962
Number of Images : 7982
X(mm) : 15.254580095
Y(mm) : 15.538067214
Width(mm) : 310.29911172
Height(mm) : 310.501818577
Scan Result : Success
SampleName : 300mm_Wafer
Alignment Angle : 0.789400
Alignment File : \\DESKTOP\Alignments\300mm_Wafer_Alignment.csv
Autofocus Set : 300mm_Wafer_AF
Autofocus Type : Automatic Predictive
Autoloader Set : None
CenterX : 170384.977373
CenterY : 170668.930430
DiePitchX : 64498.265301
DiePitchY : 90001.686035
Edge Exclude : 3000.000000
Exposure Time : 1
Filter Slider Status : None
FlatOffsetAngle : 0.000000
FlatSegmentLength : 0.000000
Focus Point Pattern : 300mm 9 Point
Focus Predictor : PreFoc Polynomial Standard
Golden Tile Number of Devices : 8
Golden Tile Scan : 1
Golden Tile Tiles per Device : 88
Illumination : Bright Field
Image Height Microns : 7410.389169
Image Height Pixels : 4268
Image Width Microns : 7888.396686
Image Width Pixels : 4398
Initiator : Manual
Install Build Version : 0.24.2.0
JobName : 300mm_Wafer_Demo_Job
JobUpdateTime : 2024-09-08 23:42:31.428
KLARFMode : 1
Layout File : D:\Scans\300mm_Wafer\Scan_006\Layout.csv
Light Intensity Control 1 : Setpoint
Light Intensity Control 2 : Setpoint
Light Intensity Control Value 1 : 0
Light Intensity Control Value 2 : 1000
Light Source 1 : Nano:Transmitted
Light Source 2 : Nano:Reflected
Mosaic Name : 300mm_Wafer_Mosaic.png
Objective : 2.5X - CFI L Plan 2.5X
OriginOffsetX : 0.000000
OriginOffsetY : 0.000000
Pattern : WholeStage
Pattern Type : 2
Radius : 149988.238372
SampleCenterX : 172401.665016
SampleCenterY : 172685.624342
SampleID : 300mm_Wafer
SampleLength : 300.000000
Scan Height Microns : 309401.818577
Scan Min X Microns : 16256.183319
Scan Min Y Microns : 16348.184435
Scan Tilt Angle : 0.000000
Scan Width Microns : 308409.111720
Stage Height Microns : 343179.700000
Stage Width Microns : 344681.400000
XOffset : -17537.000000
YOffset : -18344.000000
Analyzer
--------
StartTime : 2024-09-09 17:20:57.937
EndTime : 2024-09-09 17:21:31.650
Defects : 23189
AnalyzerName : nrad_rangeselect
Devices
-------
DeviceID : 1
X : 28645.193836
Y : 117102.611014
width : 63391.820591
height : 77005.699667
Images
------
ImageID : 2001
X : 28642.416191
Y : 117095.278032
width : 4461
height : 4169
Defects
-------
DefectID X Y W H Area
ImageID : 2002
X : 36569.712877
Y : 117096.278032
width : 4461
height : 4169
Defects
-------
DefectID X Y W H Area
ImageID : 2003
X : 44498.009564
Y : 117096.278032
width : 4461
height : 4169
Defects
-------
DefectID X Y W H Area
ImageID : 5750
X : 138607.349836
Y : 82548.104671
width : 4461
height : 4169
Defects
-------
DefectID X Y W H Area
97734 2134.8 2252.4 5.6254 3.9378 22.468003
97735 1714.6 2124.7 6.0588 3.3427 20.88575
97736 534.98 2172.5 166.51 102.38 9194.793
97737 895 2134.8 32.057 1.636 24.050257
97738 2349.7 2105 7.5578 8.3532 54.429529
97739 547.35 2070.7 4.7733 4.7733 21.518651
97740 895 2040.9 1.1251 37.127 24.050257
97741 534.98 2045.9 166.51 93.381 6940.3979
97742 1630.8 1974.5 1.1251 50.628 23.100905
97743 296.46 1921.1 7.16 11.933 74.365926
97744 895 1911.5 1.1251 50.628 35.126033
97745 534.41 1919.4 165.95 101.82 9144.4774
97746 895 1845.1 1.1251 57.941 33.54378
97747 232.89 1797.9 5.0629 6.1879 31.328624
97748 1630.8 1856.4 1.1251 125.45 58.543389
97749 534.41 1793.4 165.95 92.819 9597.6348
97750 2462.8 1754.6 24.263 9.5844 164.23794Basic Pieces of Syntax
We only add new syntax to the {fmt} library for format-specifiers; these are the parts of a {variable_name:format_spec} to the right of the first colon. There are a couple of basic ideas:
Joining format-specifiers
Instead of using a single block of characters from a : to a } to denote how to format some typed thing, we will allow a sequence of these to iteratively turn the thing into progressively easier-to-format things. This is not required in regular f-strings, which never permit a : to appear in a format-specifier.
Iterables
If you have some iterable variable containing things of type T, then it is formatted with a joiner to become something that is formatted with a format-specifier for something of type T. So for example, if we had scan_ids,was a list {1, 2, 3}, and it was formatted with {scan_ids:, :03}, that would produce the string 001, 002, 003.
Note that this is not quite the same as the range-based syntax extension described in the library, because we can add joiners.
Function calls
In some places, we embed something that should be read as a function call. Function calls are a name matching [A-Za-z_][A-Za-z_0-9]*, followed by a {}-wrapped, ,-delimited sequence of arguments. Whitespace before and after is ignored. For example:

Would require foo and bar to be variables in the surrounding context, foo to understand how to find a function bar, and implicitly that function must return a floating point number to be correctly formatted by the regular format specifier:<6.5g.
Escaped f-strings
Anywhere in a format specifier that requires a string, we can recursively have an f-string. This is done with the reserved functions f and e. The basic syntax for this is to wrap in "" and {}:
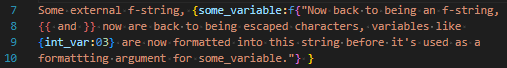
Note, critically, the space after the magenta-highlighted }. Some trailing whitespace after this function call is mandatory for distinguishing closing two scopes and intending to embed a raw } into the embedded f-string.
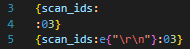
The function e has the same semantics, but additionally unescapes C-style \<char> characters. The main ones of use are \t, \r, \n, \0 (tab, carriage return, line feed, null). All but a null can technically be embedded in a string without escaping, but they can be hard to read. The following two lines express the same intent, but the latter is much clearer:
For these embeddede and fstrings, whitespace between the { and " and " and } can be freely added. If whitespace between the { and " matches exactly with whitespace at the beginning or end of the quoted string, it is removed; this allows for clean syntax separating the management of a block of text to be embedded and the text to be embedded. For example:
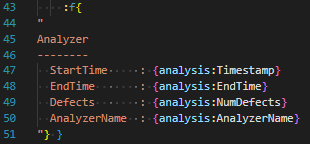
Embeds an f-string which starts with A and ends immediately after the AnalyzerName, without any trailing newline (for now, don’t worry about what theanalysis variable is).
SQL formatting
Formatting a DB
For now, a SqlDB exposes exactly one callable function, sql{row_name, sql_string}, which runs the query in sql_string and returns an iterable of rows, each of which will be exposed to subsequent formatting as a variable named row_name. For now, this means that to produce anything useful, the row must be formatted with some embedded f-string, since it is named.
Currently we are in the context of file exports for an analysis, there’s only one natural DB (the scanDB), which is exposed as a variable named db. As in other Exporters, the .njson file may have a number of queries to produce named views or other iterable rows that were returned by a SQL query.
Formatting a SQL row
Rows are dictionaries, indexed by their column titles. So something like:

Will produce a list of the ScanIDs for every scan in the Scans table, separated with", ".
If a query is embedded in the .njson file, e.g. as
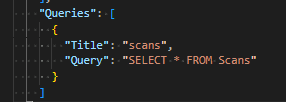
then the entire iterable of results will be named scans, and so can be formatted like:
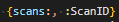
This is structurally less flexible, but is simpler syntax in the .nfmtfile.
Note that in either case, the format specifier here is simply producing some integer that is then default formatted; if this needed to be controlled with a further specifier for width or padding that occurs exactly as for any other integer.
